My latest publication [1] describes the process followed in developing the W3C Health Care and Life Sciences Interest Group (HCLSIG) community profile for dataset descriptions which was published last year. The diagram below provides a summary of the data model for describing datasets which covers 61 metadata terms drawn from 18 vocabularies. [1] M. Dumontier, A. […]
My latest publication [1] describes the process followed in developing the W3C Health Care and Life Sciences Interest Group (HCLSIG) community profile for dataset descriptions which was published last year. The diagram below provides a summary of the data model for describing datasets which covers 61 metadata terms drawn from 18 vocabularies.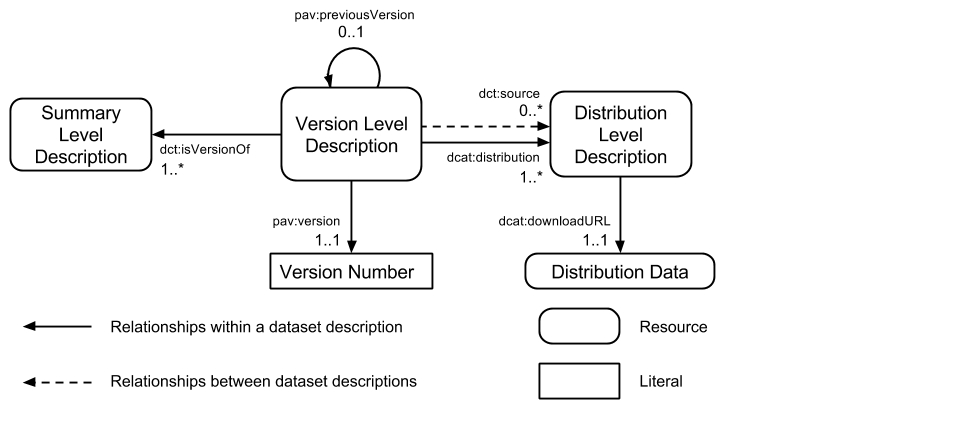
[1]
![[doi]](http://www.macs.hw.ac.uk/~ajg33/wp-content/plugins/papercite/img/external.png)
M. Dumontier, A. J. G. Gray, S. M. Marshall, V. Alexiev, P. Ansell, G. Bader, J. Baran, J. T. Bolleman, A. Callahan, J. Cruz-Toledo, P. Gaudet, E. A. Gombocz, A. N. Gonzalez-Beltran, P. Groth, M. Haendel, M. Ito, S. Jupp, N. Juty, T. Katayama, N. Kobayashi, K. Krishnaswami, C. Laibe, N. {Le Novère}, S. Lin, J. Malone, M. Miller, C. J. Mungall, L. Rietveld, S. M. Wimalaratne, and A. Yamaguchi, “The health care and life sciences community profile for dataset descriptions,”
PeerJ, vol. 4, p. e2331, 2016.
[Bibtex]
@article{Dumontier2016HCLS,
abstract = {Access to consistent, high-quality metadata is critical to finding, understanding, and reusing scientific data. However, while there are many relevant vocabularies for the annotation of a dataset, none sufficiently captures all the necessary metadata. This prevents uniform indexing and querying of dataset repositories. Towards providing a practical guide for producing a high quality description of biomedical datasets, the {W3C} Semantic Web for Health Care and the Life Sciences Interest Group ({HCLSIG}) identified Resource Description Framework ({RDF}) vocabularies that could be used to specify common metadata elements and their value sets. The resulting guideline covers elements of description, identification, attribution, versioning, provenance, and content summarization. This guideline reuses existing vocabularies, and is intended to meet key functional requirements including indexing, discovery, exchange, query, and retrieval of datasets, thereby enabling the publication of {FAIR} data. The resulting metadata profile is generic and could be used by other domains with an interest in providing machine readable descriptions of versioned datasets.},
author = {Dumontier, Michel and Gray, Alasdair J.G. and Marshall, M Scott and Alexiev, Vladimir and Ansell, Peter and Bader, Gary and Baran, Joachim and Bolleman, Jerven T and Callahan, Alison and Cruz-Toledo, Jos{'{e}} and Gaudet, Pascale and Gombocz, Erich A and Gonzalez-Beltran, Alejandra N. and Groth, Paul and Haendel, Melissa and Ito, Maori and Jupp, Simon and Juty, Nick and Katayama, Toshiaki and Kobayashi, Norio and Krishnaswami, Kalpana and Laibe, Camille and {Le Nov{`{e}}re}, Nicolas and Lin, Simon and Malone, James and Miller, Michael and Mungall, Christopher J and Rietveld, Laurens and Wimalaratne, Sarala M and Yamaguchi, Atsuko},
doi = {10.7717/peerj.2331},
issn = {2167-8359},
journal = {PeerJ},
month = aug,
title = {The health care and life sciences community profile for dataset descriptions},
volume = {4},
pages = {e2331},
year = {2016},
url = {https://peerj.com/articles/2331/}
}



