These pages apply to the XSPAN-SAEL applet.
This Help page is to introduce you to the functionality of the XSPAN-SAEL applet.
The larger screenshots on this page have been scaled down. To view the full-size image, click on the image.
The XSPAN-SAEL applet takes about 20 seconds to load. To open the Graphical User Interface (GUI) that allows browsing of the ontologies, use the "Click to Launch the XSPAN-SAEL Applet" button. The GUI opens in a new frame (window).

You are presented with a warning that there will be some delay in opening the GUI.
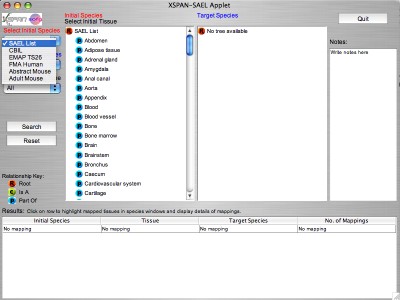
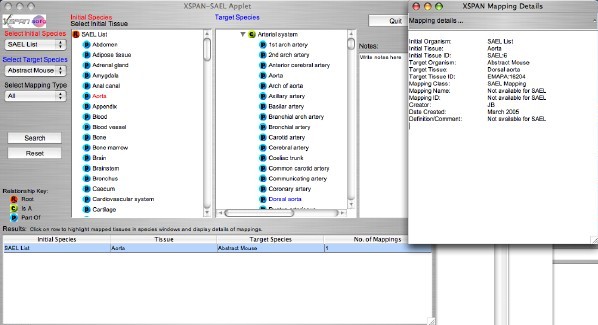
The GUI displays two ontologies side by side, and allows mappings between tissues to be identified. An initial ontology is selected from the top drop-down menu "Select Initial Species". "SAEL" is the default initial species.
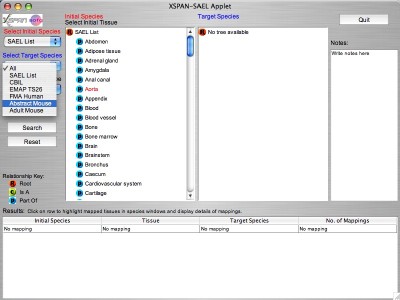
A tissue is then selected from the initial species ontology by clicking on the tree, and the selected tissue is highlighted in red ("Aorta" in the example). A target species is then selected from the second drop-down menu "Select Target Species". This species ontology then appears as the Target Species tree (Abstract Mouse" in the example). "All" is the default target species.
Click the "Search" button and a list of the mappings from the selected intial tissue to the target species appears in the table below the trees. The XSPAN-SAEL applet has only "All" as a mapping type.
Clicking on a row in the table (one for each target species) opens a new frame which displays details of the mappings, and it also highlights the target tissue/s in the target tree in blue and expands the tree to show these (may need to scroll down).
The GUI can be reset to the default settings with the "Reset" button, or another target species selected and the search repeated.
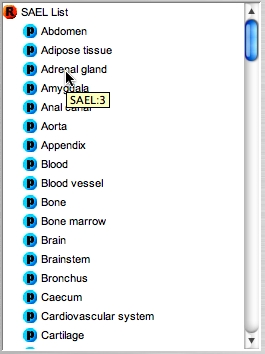
Placing the mouse cursor over a tissue in any tree will reveal the unique accession number of that tissue in the tool-tip.