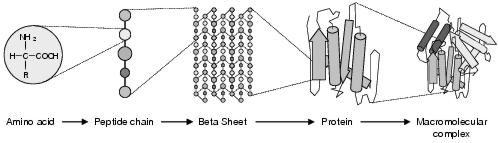
Figure 3.1: Hierarchical levels of protein structure. From left to right: amino acid, primary, secondary, tertiary, and quaternary structures.
The functioning of biological systems can be described at many levels from the interactions between individual biochemicals up through interactions at increasingly higher levels of organisation: biochemical pathways, organelles, cells, tissues, organs, organisms, populations, species, communities and ecosystems; and interactions with the abiotic environment including, for some species, cultural artefacts.
This chapter aims to show how functionality is represented at a low level within biological systems. It is divided into two sections. The first section describes the low-level components from which biological systems are constructed. The second section describes the processes by which these components are constructed and replicated. Unless otherwise indicated, this material can be found in biological textbooks such as Lodish et al. [2003], Brown and Brown [2002], and Lewin [2000].
Proteins are the main functional element of the body and play a role within almost every biological process. Specified by a sequence of amino acids, each species of protein has a unique three dimensional structure. It is this structure which determines the protein's effect upon other biological elements and, accordingly, its function within the biological system.
Amino acids are a group of molecules unified by a common structure. Of the many possible amino acids, only twenty varieties are normally found in biological systems. Each of these is distinguished by a characteristic chemical residue called the side chain which through variance in size, shape, charge, reactivity and hydrophobicity, gives each amino acid a particular chemical signature. A protein is a complex of one or more polypeptides - sequences of amino acids linked together by peptide bonds - arranged in a three-dimensional structure. This structure, termed the native state, is an attribute solely decided by the nature of the polypeptide sequences and hence, to a great extent, the chemical behaviour of each amino acid1.

Figure 3.1: Hierarchical levels of protein structure. From left to right: amino acid, primary, secondary, tertiary, and quaternary structures.
A protein's structure is specified by four hierarchical levels: the primary, secondary, tertiary and quaternary structures (see figure 3.1). Primary structure is the linear amino acid sequence(s). Secondary structure describes local organisation within the polypeptide chain caused by attractions and repulsions between chemicals in different amino acids. There are a number of basic components which may form within the chain. The term random coil refers to those parts of the chain where no specific pattern of structure emerges. a-helices and b-strands are the most common structural components within a protein. An a-helix is the result of hydrogen bonding between oxygen and hydrogen atoms along the polypeptide backbone, causing the local chain to form a stable helix. This a-helix acts like a sturdy and inflexible rod, making it a good structural member for mechanical action. Where a-helices have one side hydrophilic (soluble in water) and the other hydrophobic (insoluble in water), a state termed amphipathic, they may combine with other a-helices to form a higher-dimensional, tougher helix. b-strands are sections of polypeptide which do not coil, but remain straight and non-helical. Though fairly weak by themselves, they hydrogen bond with other b-strands (running parallel or anti-parallel) to form b-sheets. The presence of amino acid residues on both sides of the sheet can allow for interesting behaviours to emerge - for instance if those on one side are hydrophilic and the other side hydrophobic. Tougher structures result when sheets form stacks.
Turns, composed of three or four amino acids, are sharp bends in the chain resulting from hydrogen bonding between the residues at either end of the sequence. Motifs are distinctive combinations of secondary structures, with characteristic function and primary structure. An example is the zinc finger, a motif consisting of one a-helix and two b-strands. These form a cage around a single zinc atom; the assembled motif resembling, and offering similar function to, a finger. The zinc atom involved in this motif is an example of a prosthetic group - a tightly bound non-peptide molecule or metal which provides structural support, a binding site, or some other function to the protein.
Tertiary structure defines the folded form of the protein when introduced to an aqueous environment - where hydrophobic interactions pull the secondary structure into a more compact form. Typically, the protein will configure itself into a number of distinct regions, called structural domains, which encompass a section of the polypeptide chain containing numerous secondary structures. A cluster of structural domains which together provide a localised function within the protein are deemed a functional domain. Functional domains which recur in many proteins (and therefore exist as building blocks) are known as tertiary domains.
Where a protein consists of more than one polypeptide, a final layer of structure - quaternary structure - describes the positioning of polypeptide subunits. In certain circumstances, multiple proteins may form an aggregate structure. These are called macromolecular assemblies.
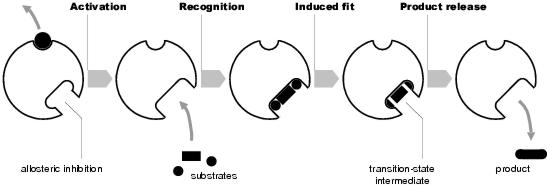
Figure 3.2: Enzyme activity
Enzymes are proteins which act to catalyse other chemical reactions. For a chemical reaction to take place, an activation energy must be met. Usually, this is provided by the kinetic energy of the reactants. However, for many reactions, the kinetic energy required is substantial and is only available at hot temperatures. In biological systems, ambient temperature is relatively low, making most reactions energetically unfavourable. To make these reactions possible, either the activation energy must be met - not usually possible - or it must be lowered. This is the function of enzymes (see figure 3.2). Enzymes possess specificity, an ability to recognise2 only certain chemicals and bind exclusively to these. The chemicals recognised, the substrates, are the reactants needed for the reaction. By bringing these together, the kinetic energy required for the reaction to occur is reduced, and hence the activation energy of the reaction is lowered.
Most important reactions do not take place immediately. Rather, the reaction progresses through a number of transition states. During this process, the substrates are converted through a series of transition-state intermediates until, after the final transition stage, they become the reaction's final products. For an enzyme to effectively catalyse a reaction, it must not only bind to substrates, but also to transition-state intermediates.
Binding of substrates occurs at active sites on the enzyme. Active sites are produced by a precise arrangement of amino acid residues which, when in contact, bond non-covalently with complementary sites on the substrate's surface. This is called the lock-and-key mechanism. Recognition of a substrate may also cause structural change in the enzyme, induced fit, which brings the substrate into closer contact with the active site. Non-covalent bonding holds the substrate in place. However, the enzyme may also form covalent bonds with a substrate. It is these bonds which change the nature of the substrate, converting it into a transition-state intermediate. Subsequent changes in the substrate occur either by contact with other reactants, or by further enzymatic action (making and breaking of covalent bonds). In order for the correct bonds to be made, amino acid residues alone may be insufficient. In these circumstances, prosthetic groups may be used. Prosthetic groups used in this way are called co-enzymes.
Since chemical reactions require energy and resources, for purposes of efficiency it is desirable that reactions only take place as and when they are required. Consequently, the function of enzymes are regulated so that reactions are only catalysed when the current chemical conditions make them useful. This is achieved through effector-binding sites which, when bound, either inhibit or activate the enzyme. The molecules which bind to these sites are called effectors. In the case where the (inhibiting) effector is the product of the reaction which the enzyme catalyses, this is called feedback inhibition. Moreover, in real biological systems, products of reactions often become reactants of other reactions. In these cases, the product of the final reaction may be the effector of the enzyme involved in the first reaction. Hence, this enzyme will only become functional when the end product is in short supply and, in effect, the entire pipeline will be disabled at other times (since the first reaction produces the reactants for the second, and so on). Inhibition and activation occur through conformational change induced by bonding at the effector site. This change of shape for purposes of regulation is called allostery.
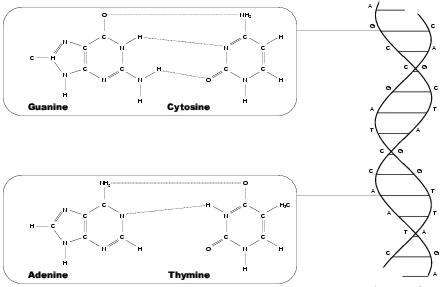
Figure 3.3: Nucleotides of DNA
While proteins provide the functional elements of a biological system, it is nucleic acids which specify and aid construction of these units. These roles are provided, respectively, by deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are polyesters composed of nucleotides. Nucleotides, like amino acids, are small molecules with common structure, differentiated between by side groups. These side groups are called bases, and are of two types - purines and pyramidines. Purines are substantially larger than pyramidines. The purines found in biological systems are Adenine and Guanine, each identified by their initial letters, A and G. The pyramidines are Cytosine (C), Thymine (T) and Uracil (U). Nucleotides with Thymine bases are only found in DNA and those with Uracil are found only in RNA. Nucleotides are formed when nucleosides, consisting of a sugar and a base and found free-floating in cellular fluids, are joined together by phosphate bonds. A nucleotide is then the aggregate of a sugar, a base and one phosphate bond. The two ends of the unit are called 5' and 3'. A 5' is always connected to a 3', and hence the entire nucleic acid has chemical polarity, running from a 5'-end at the beginning to a 3'-end at the finish.
DNA is formed from two anti-parallel polynucleotide strands, fused together by various non-covalent bonding actions. Most important of these is hydrogen bonding between bases in the two strands, forcing the amalgam into a double-helical arrangement (see figure 3.3). The linking of two bases - a purine in one strand with a pyramidine in the other - is called base-pairing. Base pairs in DNA are almost always either AT (Adenine bonded with Thymine) or GC (Guanine and Cytosine). The natural form of DNA is called B-DNA. In addition to hydrogen bonding between base pairs, the structure is stabilised by hydrophobic interaction and Van der Waals bonding3 between helical sections, or turns. The helix makes a complete turn every ten base pairs, which is about 3.5nm. However, A-form DNA - which only remains stable in non-aqueous environments - turns every eleven base pairs. This is a more compact form of DNA, with a turn of about 2.3nm, but only occurs when DNA is removed from solution. Both A- and B-DNA are right-handed varieties. A further form, Z-DNA, is left-handed, but has never been observed in natural biological systems. DNA in eukaryotes, such as animals and plants, is found in linear form. However, in prokaryotes and viruses DNA is circular, with the 5'-end attached to the 3'-end. Sometimes sections of circular DNA may become underwound. This state is energetically unfavourable, and so the entire strand is pulled towards a more favourable state - either by reducing the overall twist or by forming supercoils (where the degree of supercoiling is measured as writhe).
DNA is an information store, a purpose to which it is well-suited due to its relative long-life and stability. RNA sacrifices long-life for lability; a fact reflected in its multitude of uses within biological systems. It can occur in single-stranded and double-stranded varieties, linear or circular, can hybridise with DNA, and can combine with proteins to form ribonucleoprotein complexes. Like proteins, RNA can form three-dimensional structures: allowing expression of catalytic and auto-catalytic behaviours (for instance, breaking and splicing other RNA molecules). These structures are described by three levels of organisation. Primary structure describes the base sequence, secondary specifies two-dimensional structure such as loops and hairpins, and tertiary describes interacting two-dimensional features which form three-dimensional structures such as pseudoknots.
The purpose of DNA in biological systems is to store a description of the organism in which the DNA is found. This information, called the genome, is expressed by a language written in the genetic code - the alphabet of bases found in DNA - namely, A, C, G and T. However, this information is not a blueprint, but rather a highly decentralised developmental plan which describes, by specifying systems of proteins, how the organism will function at a local level. The overall nature of the organism is then emergent from the sum of these local functions.
According to Mendel [1965], the founder of genetic science, a gene is a unit of inheritance, a `particulate factor that passes unchanged from parent to progeny'. From a functional viewpoint, a gene is a stretch of polypeptide chain encoding one protein, or more exactly, a fragment of DNA which can be transcribed by messenger RNA.
Although the language and chemical structure of prokaryotic and eukaryotic DNAs are virtually the same, there are two major differences in genetic structure. The first of these concerns the unit of transcription. In prokaryotes, it is normal for several genes to be encompassed by the same transcription unit. This means that a single mRNA can encode several different proteins, each of which can be synthesised independently by a ribosome. In eukaryotes, by contrast, ribosomes may only begin synthesis at the beginning of an mRNA strand, entailing that eukaryotic mRNA may only transcribe from a single gene. This is called monocistronic RNA. Prokaryotic mRNA is polycistronic. The cluster of genes from which this is transcribed is an operon - with transcription starting from a short stretch of DNA called a promoter. If this promoter is mutated, then all the genes in the operon may become non-functional, a fact which makes prokaryotic DNA less fault-tolerant than its eukaryotic cousin. However, this approach is slightly more efficient than having many separate transcription and synthesis events - and efficiency is very important to a prokaryote, where evolutionary pressure selects against any waste of energy. Moreover, low-level efficiency is relatively unimportant to a eukaryotic organism, for which behavioural effectiveness is the dominant evolutionary selector. This, too, explains the second major difference between prokaryotic and eukaryotic DNA. Prokaryotic DNA is tightly packed, with almost all the polypeptide used to encode functional genes. By comparison, eukaryotic DNA consists mostly of non-coding DNA, the relative quantity of which varies widely between species and does not correlate with the size or complexity of the organism. Eukaryotic genes consist of exons, coding segments, and introns, non-coding segments. During transcription, both coding and non-coding parts are copied to mRNA. Before protein synthesis, mRNA excises its non-coding introns to form a continuous stretch of coding RNA.
Solitary genes are genes which occur only once in the genome, accounting for between twenty-five and fifty percent of all genes. A gene family is a set of nearby4 genes which encode similar, but not identical, amino acid sequences (a protein family). Polypeptide sequences which are similar to genes but are non-functional are called pseudogenes. Quite often, sequences of bases occur over and over again in a repeated array. Depending on whether the sequence is a gene or is non-coding, these are called either tandemly repeated genes or repetitious DNA fragments. Tandemly repeated genes encode proteins for which demand is greater than that which can possibly be transcribed from a single gene in a given time period. To meet demand, transcription of many identical genes occurs in parallel.
More generally, there are three classes of eukaryotic DNA. These are identified, and named, according to how fast they re-associate after their strands have been separated. Where there are tandem arrays of short sequences (5-10 base pairs), sections of one strand can bond to many sections on the other, pulling the strands together very quickly. Such DNA is called rapid reassociation rate, or simple sequence, DNA. Due to the way it forms bands around other DNA when centrifuged, it is also known as satellite DNA. Regions of fewer repeats are minisatellites, with small differences in the length of minisatellites between members of the same species providing the basis for genetic fingerprinting. A single variety of simple sequence DNA can occupy up to one percent of the genome in total, and is often found in specific areas of the chromosome.
Intermediate reassociation rate, or intermediate repeat, DNA represents many occurrences of larger base sequences. Compared to simple sequence DNA, there are relatively few varieties of these, although each variety occurs in large numbers. Repeating sequences of between 150 and 300 base pairs are classed as short interspersed elements, SINES, whereas those of 5000 to 7000 base pairs are LINES, long interspersed elements. Intermediate repeat DNA is either found in large tandem arrays (e.g. functional gene tandem arrays) or scattered randomly around the genome. This latter class includes mobile DNA elements - DNA sequences that are able to move or copy themselves to other regions of DNA. These sequences, which have no real purpose other than self-replication5, occupy about thirty percent of the human genome. Slow reassociation rate DNA, or single copy DNA, occupies between fifty and sixty percent of the genome. Of this, only five percent encodes genes - the rest being spacer DNA with mostly no known function.
Although seen as a molecular parasite, using the organism's transcription facility without giving anything in return, mobile DNA is thought to have evolutionary significance. On the whole, transposition of mobile DNA is balanced by mutation, which destroys existing copies with no disadvantage to the organism. However, it does lead to variance in the lengths of chromosomes - meaning that it is possible that two chromosomes of unequal length will be crossed over. The result of this is unequal crossover, which can lead to duplication and mutation of existing genes. Mobile DNA can also carry with it parts of genes it has overwritten. If these parts are then copied into other genes when the mobile DNA moves, exon shuffling takes place - the creation of novel genes from combinations of pre-existing exons [Gilbert, 1978].
There are two broad classes of mobile DNA, categorised by their transposition mechanism. Transposons remain as DNA throughout the move. They are either excised or copied from their original location and then inserted at their new location. Retrotransposons transpose via an RNA intermediate, and hence are always copied rather than moved. RNA polymerase encodes the retrotransposon as RNA. An enzyme called reverse transcripterase then copies this to a new segment of DNA which is then inserted into the chromosome. Retrotransposons are either viral or non-viral, with viral retrotransposons encoding a viral shell which, when synthesised, allows the retrotransposon to leave the cell and infect other cells and organisms. Non-viral retrotransposons are either LINES or SINES. LINES encode reverse transcripterase. SINES, however, use the reverse transcripterase synthesised by LINES, meaning they can be much shorter (in effect, they are hyperparasites).
Prokaryotes also contain selfish DNA. In bacteria, insertion sequences (IS elements) are 1.5kb segments of single-strand DNA which invade normal double stranded DNA (homoduplex), forming a heteroduplex with one strand containing the extra IS element. Insertion sequences include instructions for synthesising transposase, which allows the IS element to move within the bacterial DNA. However, since bacterial DNA is tightly packed, these moves are likely to generate fatal mutations. For this reason, surviving IS elements transpose very rarely.
The introduction of new, useful, genes as well as occasional beneficial effects of mutation and exon shuffling are rare chance events. The role of mobile DNA is, on the whole, non-functional. By contrast, local cellular processes sometimes carry out rearrangements of DNA with a specific functional intent. These include inversion of DNA sequences, gene conversion, amplification and segment deletion. The role of inversion is varied, and depends upon the organism. For instance, in the bacterium salmonella, it is used to alter the expression of certain surface proteins. Once the host organism has produced antibodies for the primary infection, bacteria which experience this inversion will not be recognised, producing a secondary infection for the body to combat. Gene conversion results in a component of an active gene being updated with an inactive part from elsewhere in the chromosome, changing the protein produced by the gene. The uses of this mechanism, again, are varied. Gene amplification, also called polytenation, causes parts of chromosomes to be replicated. The replicants are either than released, or remain connected. This is the mechanism used to produce lots of copies of the rRNA gene. It is a dynamic alternative to tandem arrays. Finally, segment deletion is involved in the separation and rearrangment of segments of DNA, allowing many forms to be generated from a set of DNA segments (e.g. antibodies).
In many organisms, genomic DNA is arranged into chromosomes. Dividing the genome into storage units in this manner makes the large amount of data easier to store and handle. However, the genome is very large. In its natural form, even split into sub-units, the DNA is far too voluminous to fit into a single cell, let alone a nucleus. In prokaryotes, which have relatively small genomes, the natural random coil formed by the single circular chromosome would be one thousand times larger than a bacterial cell. Much of this expanse is due to electric charge repulsion between the negatively charged phosphate groups in the DNA backbone. The prokaryotic solution to this problem is to introduce positively charged polyamine groups, which associate with the phosphate groups to shield the charges. Additionally, numerous small proteins bind to the DNA, folding it into a more compact structure, and finally, special enzymes induce supercoiling.
The eukaryotic solution to the compaction problem is called chromatin, a complex of DNA and structural proteins. These proteins, H1, H2A, H2B, H3 and H4 are all members of the histone family and retain a high degree of similarity between species. Two each of H2A, H2B, H3 and H4 form a histone octomer, a roughly cylindrical shaped object that acts like a reel around which DNA can be wound. A nucleosome is a histone octomer with slighty less than two windings of DNA (146 bases). This is the primary structural unit of chromatin. Further organisation produces a solenoid form, euchromatin, with six nucleosomes per turn. This structure is stabilised by an association of histone H1 with each nucleosome - forming chromatosomes. During interphase (the period between cell divisions), euchromatin is the normal level of chromosome structure, the non-condensed form of chromatin. During metaphase (cell division), when chromosomes become visible to the light microscope, the chromosome experiences several degrees of supercoiling, forming a compact structure called heterochromatin, the condensed form. This is supported by an internal scaffold6 of non-histone proteins. The number, size and shape of heterochromatin complexes during metaphase is an organism's karyotype. During interphase, some sections of the chromosome remain as heterochromatin, occuring in regions where no transcription takes place. When interphase chromosomes are stained by certain dyes, these regions of heterochromatin form dark bands. Staining of metaphase chromosomes also produces characteristic bands. However, the cause of this banding is unknown, yet provides a useful roadmap within the organism's karyotype.
Most sequences of bases in DNA are either protein encoding genes or non-coding redundant regions. However, other sequences are designed with a functional role. These have arrangements of bases which are recognised by various types of binding proteins, such as transcription sites. Three such regions are of particular importance for chromosome duplication and segregation - autonomously replicating sequences (ARSs), the centromere and the telomeres. ARSs are origins of replication. These are bound to by special proteins which start the replication process. Centromeres have binding sequences for proteins which hold and follow the microtubule spindle. Telomeres7 protect the ends of chromosomes, and are placed after replication. They also provide a binding point outside of the main chromosome where replication of the chromosome ends can occur from. Otherwise, it would not be possible to replicate the last few bases of a linear chromosome, creating shorter chromosomes each generation.
Cells are the fundamental unit of most biological systems (or the unit, in the case of some). It is local processing done by cells which, when combined with communication between cells, lead to the emergent behaviour of a single organism. This local processing, called the metabolism, is in turn the sum of all chemical processes which occur within the cell.
Within the biological community, the prevailing view is of a single heritage to all cells. However, from the current standpoint in evolution, cells can be seen as falling into two broad categories - prokaryotic and eukaryotic. Organisms which consist of these types of cell are called, respectively, prokaryotes and eukaryotes. Prokaryotes encompass the most primitive organisms on Earth, the bacteria. This cell lineage can be further divided into eubacteria and archaebacteria, the latter of which is considered by some as a separate lineage to the prokaryotes. Eubacteria are common, and consequently well studied. Archaebacteria live in unusual environments such as swamps and sulphur springs, and at the cellular level have many features in common with eukaryotes. Eukaryotes include animals, plants and fungi.
Both prokaryotic and euakaryotic cells are contained within a plasma membrane. The plasma membrane is a semi-permeable layer which maintains the integrity of its contents by controlling inward and outward movement of chemicals. Prokaryotic cells are fairly unstructured. Eukaryotic cells, however, contain extensive internal membranes which organise the cell into compartments of local metabolic processing, organelles. The largest of these, and the structure which gives the eukaryotic cell its name, is the nucleus, the location of the cell's DNA. However, exactly which organelles are found in a cell depends upon the cell's type - which is likely to be decided by the surrounding tissue. In fact, not all eukaryotic cells have a nucleus. The red blood cell, for example, has, and needs, very little by way of internal structure since it does not divide and does not communicate with other cells. Most animal cells, however, contain at least a nucleus; mitochondria, to provide energy; centrioles, which are used in cell division; and lysosomes, which handle recycling. Plant cells have similar components, but use chloroplasts rather than mitochondria to provide energy, and often contain vacuoles for storage of nutrients and fluids. The region of the cell outside the organelles is termed the cytosol, where much of the cellular metabolism takes place. Until fairly recently, this space was assumed to be fairly unstructured. However, higher resolution microscopes have shown the cytosol to contain a filamentous cytoskeleton which defines a grid-like structure within the cell and holds the organelles in place. It is theorised that the proteins and enzymes within the cytosol are also well organised spatially, possibly by binding to particular fibres in the cytoskeleton.
Most important cellular functions are anabolic, requiring complex molecules to be assembled from simpler ones - and since these reactions are energetically unfavourable, energy must be provided for them to occur. For most reactions, this is provided by a chemical called adenosine 5'-triphosphate (ATP) which, when catabolised, releases energy which may then be used in a nearby anabolic reaction. Cells do not have direct access to ATP from their environment, but must rely on some other form of energy as a source. For example, animals obtain energy from food in the form of sugars which are eventually converted to glucose and distributed to all cells of the body via the blood stream. Plants, on the other hand, rely upon light energy. The conversion of these energy sources into ATP is handled by organelles within the cell. In animal cells, this function is provided by mitochondria, large organelles covering up to a third of the cellular space in total. Mitochondria have their own DNA8, used to synthesise proteins (ATP synthase) which convert large glucose molecules into many small, easy to use, ATP molecules. This occurs on the surface of a large folded inner membrane. Plants have similar organelles called chloroplasts. These contain DNA used to synthesise pigment (chlorophyl) and enzymes which, together, convert light into ATP.
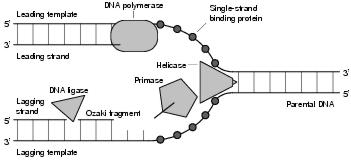
Figure 3.4: DNA Replication
The process of DNA replication is said to be semiconservative. From two existing complementary strands, two new strands are synthesised (see figure 3.4). This produces two daughter molecules, each consisting of one old strand and one new. Synthesis of a new strand starts at a replication origin, of which there may be many, somewhere within the existing strand. From this point, the replication process may be either omnidirectional (moving one way along the strand, but not the other) or bidirectional. The former occurs in some bacteria, a circular genome meaning that replication will eventually reach the other end even if only one replication origin is involved. Bidirectional replication is normal for eukaryotes with linear genomes. At the start of replication, the two existing strands are still connected together, with disconnection occurring incrementally at the same time as replication. This means that synthesis of both strands can take place at the same place at the same time, producing something that looks like a bubble. In some viruses this is not the case, and synthesis of the two strands begins in different places.
The region of DNA served by a single replication origin is a replicon. The current point where the strands are separated up to, and new DNA is being synthesised, is called a growing fork. For bidirectional replication, there are two of these. The region between the forks, or between the fork and the origin, is called a replication bubble. In humans, replication forks move at about one hundred base pairs per second. The entire genome is about 3x109 base pairs, divided into between 10,000 and 100,000 replicons. Replication of the entire genome takes about eight hours, with redundancy in the number of replication origins meaning replicons need not be active throughout this entire period.
A growing fork moves in one direction along the existing DNA. However, DNA can only be grown in one direction, from the 3'-end to the 5'-end. Since the strands in DNA are anti-parallel, and both strands will be copied at the growing fork, only one of the new strands (the leading strand) can be grown throughout from 3' to 5'. The strategy used for the other new strand (the lagging strand) is to grow small fragments of DNA as groups of bases become available. These Ozaki fragments are then joined together to form a complete strand.
Replication origins (ARSs) consist of tandemly repeated short sequences which are recognised by multimeric origin-binding proteins. These proteins bootstrap the replication process by assembling the replication enzymes at the right locations. The first of these enzymes detaches the two strands at the replication point, forming a small gap called an open complex. Helicase then binds to this open complex, taking on the role of strand separator at the leading edge of the growing fork. As the strands are separated, small molecules - single-strand binding proteins (SSBs) - bind to the exposed strands, preventing the DNA from re-annealing. Primase, working to the rear of helicase (and forming a macromolecular complex called a primosome), is responsible for creating new RNA primers as necessary (only once on the leading strand, and many times on the lagging strand). DNA polymerase III uses these RNA primers, growing new complementary DNA bases according to the exposed strands, whilst removing SSBs. DNA polymerase I removes the RNA primers from DNA strands and DNA ligase joins DNA fragments together. In prokaryotes, DNA is particularly amenable to supercoiling, making it difficult to separate and process individual strands. The solution to this is an enzyme called topoisomerase, which removes supercoils at the growing fork and re-establishes them later.
During synthesis, it is possible that errors may occur. To combat this, DNA polymerase offers a proofreading capability which inspects the newly synthesised strand as it forms. If an error is found, the incorrect section of the new strand is removed and the synthesis action of polymerase is reactivated to update the strand with the correct information. More generally, environmental influences may cause errors to occur in DNA at any point of its lifetime. The damage caused may be missing, altered or invalid bases, bulges, bonding between side chains, strand breakage, strand cross-linking or fragmentation. The damage is either repaired directly, or more commonly, an excision-repair mechanism removes the damaged section and updates it using DNA polymerase. Some researchers also hypothesise that DNA sequences contain error correcting codes which minimise the impact of the errors that are not repaired by the excision-repair mechanism [Battail, 2003].
Transcription is the synthesis of a messenger RNA strand from a DNA template. Transcription is similar to DNA replication in that RNA polymerase recognises start sequences in the genetic code, binds and reads DNA, and constructs complementary RNA sequences. The process is similar in both prokaryotes and eukaryotes, although substantially more complicated in the latter.
Prokaryotic genes occur in conserved structures known as operons, genetic units that tie groups of genes into a single transcription event. An operon is a tightly packed polypeptide sequence consisting of a control region followed by a group of genes. The control region consists of a promoter, where RNA polymerase first binds, and an operator, which enables or disables transcription of the operon.
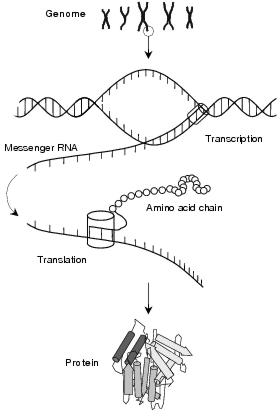
Figure 3.5: Protein synthesis through transcription and translation
Promoter sequences contain distinctive base-sequence patterns which are recognised by initiation units within RNA polymerases. There are a number of different types of RNA polymerase, and each binds to a particular pattern. The strength with which an RNA polymerase binds, and therefore the frequency at which an operon is transcribed, depends upon how accurately a promoter implements this pattern. Hence, genes which should be transcribed often have very accurate, or strong, promoters whereas those which are seldom transcribed have only a passing resemblance to the ideal pattern. For most operons, the important promoter sequences lie around ten and thirty-five bases downstream of the genes. The parts of the RNA polymerase that bind to these regions are collectively called the initiation factor, or s. The complex of subunits that continue transcription after binding form the core polymerase. Together, the intitiation factor and the core polymerase form a holoenzyme, a complete and functional molecule.
Operon expression is controlled by molecules (usually enzymes) called regulators that, through binding to control locations, turn transcription on or off. Positive regulators are called activators. By binding to both promoter sequences and RNA polymerase, these strengthen the hold between the two, making transcription more likely. Hence, activators make a promoter stronger. Repressors, negative regulators, do not effect the promoter but rather bind to the operator region, blocking RNA polymerase and preventing transcription. Operators typically consist of an inverted repeat sequence (a sequence followed by its repeat in reverse), each repeat forming a half-site. Repressors are dimeric, consisting of two units, each unit binding non-covalently to a different half-site - the bond made stable by a-helices which the repressor inserts into major grooves of the DNA helix. A set of operons controlled by the same regulator is called a regulon. Each member of the regulon may have a different affinity for the regulator, in the same way that different promoters have differing affinities for a certain RNA polymerase. Certain molecules, by binding to recognition sites on the repressor, may cause the positions of its binding a-helices to change, deactivating the repressor. This allows repression to be enabled or disabled according to the chemical environment. Hence, gene expression is dynamic, changing according to the products of other genes and the current needs of the molecular system.
Not all prokaryotic genes follow the schemes outlined above. In particular, some genes are transcribed by non-standard RNA polymerases with altered s-factors. These bind at many different sites, some within the normal promoter region and others in more distant places. Also, some RNA polymerases cannot use ATP directly, requiring the help of protein intermediaries. These intermediaries also bind to the DNA, often quite a distance from the operon being transcribed. Although the exception for prokaryotes, where much gene regulation is simply metabolic response to the presence of nutrients, these more complicated systems are the norm for eukaryotes. Here, complexity reflects the fact that genes, cells, tissues and organs all have highly-linked actions and responses, meaning that the influences on gene expression may be far from simple.
In eukaryotes, genes are transcribed on an individual basis rather than arranged into operons. RNA polymerase II is responsible for the transcription of protein-encoding genes, whereas polymerases I and III operate solely upon RNA-template genes. Eurakyotic RNA polymerases do not contain an initiation factor or carry out ATP-hydrolysis, relying upon DNA-binding proteins to provide these facilities. This results in a very flexible process, with some initiation complexes consisting of many large protein complexes. These factors bind to DNA promoters at various locations, and through sequential interactions, guide the activity of the RNA polymerase. Eukaryotic promoters are of several types. The strongest are TATA-boxes and initiators, similar in structure to prokaryotic promoters. An interesting exception is the CpG (where `p' denotes the phosphate bond) island, a CG repeating sequence which occurs just upstream of the first exon. The sequence CG is statistically under-represented in the eukaryotic genome, and hence stretches are easily recognised as promoters and unlikely to be confused with other genetic information9.
Promoter sites are found anywhere between tens and tens of thousands of base pairs away from exon sites. Promoter sites at a distance of less than two hundred base pairs are called promoter-proximal elements, and those at a greater distance, enhancers. Enhancers can occur upstream of exons, within introns, or even downstream of the entire gene. Typically, multiple activator proteins will bind to a single enhancer site, sometimes forming heterodimer complexes with members of the same class. Activators are often modular, with distinctive (even separable) DNA-binding and activation domains. Repressors are less-well understood than activators, yet appear to have a similar structure and capability to regulate from distant binding sites10.
The purpose of transcription regulation in eukaryotes is different to prokaryotes. Prokaryotes are single-celled and chiefly concerned with metabolising a variety of nutrients. Eukaryotes, on the other hand, are multi-cellular. Transcription regulation is the mechanism used to make cells specialised, with the pattern of gene expression in a cell reflecting its lineage and hence its type with respect to the surrounding tissues. Regulators are inherited from the cytosol of its parent cell, and these, through transcription regulation, decide the format of the cell's metabolitic activity. Other regulators are universal, passing configuration signals throughout the organism. These either act directly, by binding to promoter regions, or indirectly, through signal transduction pathways. For example, lipid-soluble hormones pass through cell walls and excite receptors which then release inhibitors which block the action of transcription factors. Furthermore, lipid-insoluble hormones excite receptors on cell surfaces, which then transmit signals through the plasma membrane to excite receptors within the cell etc etc. In practice, far more complicated pathways exist (see section 4.2 for a full discussion of signalling pathways).
Transcription in eukaryotes is made more complex by the structural organisation of chromatin. Areas of heterochromatin, which are very compact, are inaccessible to transcription factors. This entails that transcription cannot take place during cell-division. Furthermore, certain sections of chromatin remain condensed during interphase. These areas are not transcribed - even though theoretically-active genes exist within them. Some of these areas have become unused over evolutionary time, making it more efficient for them to remain compressed. Other areas are only found compressed in certain cells. This form of regulation is equivalent to regulator molecules in the cytosol, making the cell specialised. Human females inherit two X-chromosomes from their parents. Early on in development, one of these chromosomes becomes, and remains, condensed. Hence, only one of the human X-chromosomes, decided randomly, is transcribed in a given human.
Finally, mitochondria and chloroplasts have gene transcription systems similar to prokaryotes. Both these organelles have small genomes encoding a small number of proteins. Mitochondria, in particular, have very simple transcription systems - with mitochondrial RNA polymerase being the simplest protein that can carry out all stages of transcription.
For prokaryotes, cell-division and reproduction11 are typically the same thing. The circular DNA within the cell becomes attached to the cell membrane. The DNA replicates, separates, and the new strand becomes attached to the plasma membrane at a different, but nearby, point. The membrane between the two attachment points and the membrane on the opposite side of the cell then move together. When they join, the cell separates into two daughter cells - each with its own DNA and a fifty percent share of the original cytosol.
The cell cycle for eukaryotes is somewhat more complicated and can be expressed by two different processes, mitosis and meiosis. Mitosis is normal cell division, accounting for regeneration and growth and producing daughter cells that are genetically identical. Meiosis is the production of germ cells (eggs and sperm) to meet reproductive needs, generating daughter cells that are genetically different.
The mitotic cell cycle consists of four phases, illustrated in figure 3.6. At the beginning is a gap of inactivity, during which the cell goes about its normal metabolic functions. At some point in time, depending upon the type of cell and how frequently it should divide, the cell enters the synthesis phase - during which nuclear DNA is replicated to produce two identical sets. Following from this is another, shorter, gap of inactivity, concluded by the mitotic phase. The mitotic phase, in turn, is carried out in four stages.
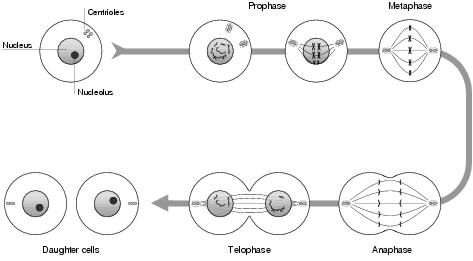
Figure 3.6: Mitotic cell division
In eukaryotes, it is normal for nuclear DNA to be split into a number of units, each contained within its own macromolecular complex. This complex is called a chromosome and consists of a single molecule of DNA bonded to various proteins and structural members. In sexual species, there are two sets of chromosomes - the maternal and the paternal - inherited, respectively, from the organism's mother and father. After the synthesis stage of mitosis - where chromosomes are duplicated - each chromosome remains attached to its duplicate, forming a structure called a sister chromatid. Since this is the only time when chromosomes become easily visible in a microscope, this X-shaped form has become the standard representation of a chromosome - even though it really represents two chromosomes.
So, before the mitotic phase, the cell has twice the normal quantity of DNA, arranged into sister chromatids in the nucleus. The first stage of mitosis is prophase, during which the centriole organelle replicates - each centriole moving to opposite ends of the cell. Between the two centrioles, and spanning the length of the cell, microtubule spindles develop, each attaching itself to the middle of a sister chromatid and providing tracks along which the chromatids may move.
Metaphase, the second stage of mitosis, is characterised by the disappearance of a distinct nucleus. By the end of metaphase, the sister chromatids have become aligned along the cell's equatorial plane. This is followed by anaphase. Here, the sister chromatids become detached, each moving in a different direction along the microtubule spindles. Finally, in telophase, cytokinesis - division of the cell's cytoplasm - occurs, chromosomes group together, and nuclei reform in each of the daughter cells. During cytokinesis, mitochondria or cytoplasts, which have their own cycle of prokaryote-like division, are shared roughly equally between the daughter cells. Both daughter cells are now roughly identical, containing all the important organelles and a normal quantity of DNA.
Meiosis is the production of sex (or germ) cells; cells which contain half the chromosomes of their parent (stem) cell. During reproduction, germ cells from the mother and father organisms join to form a zygote, a cell containing a full set of chromosomes which define the genome of a new organism.
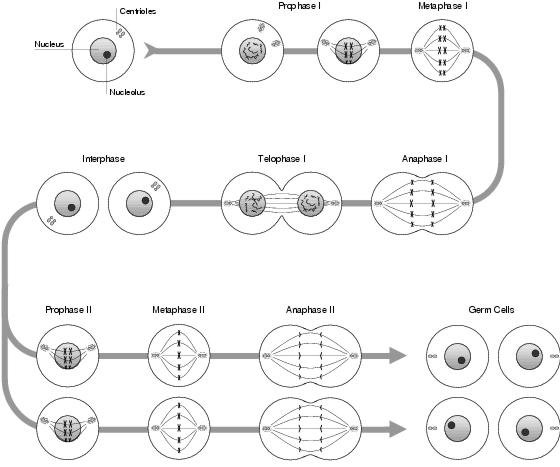
Figure 3.7: Meiosis
The mechanism of meiosis (see figure 3.7) is similar to mitosis. Up until early metaphase (where sister chromatids have been formed and the nucleus has denatured), the process is identical. However, instead of the chromatids aligning linearly along the equator, they line up in homologous pairs; with each maternal sister chromatid lying next to its corresponding paternal sister chromatid. Each sister chromatid in this pair will move, non-deterministically, to a different daughter cell during anaphase; and following this chromosome segregation, each daughter cell will contain exactly one of each chromosome type.
During late metaphase, and before chromosome segregation takes place, the aligned chromatids experience recombination: during which genetic material is transferred between the chromatids. Recombination involves a process called crossover: the exchanging of sections of DNA between the chromosomes; and although not explicitly transferring genes, this is the most likely outcome. The mechanism of crossover is explained in the following section. Following recombination, within anaphase and telophase, each homologue of a sister chromatid pair moves towards a different end of the cell and cytokinesis occurs. Unlike in mitosis, the sister chromatids do not split into separate chromosomes. The result is two daughter cells, each containing the normal amount of DNA, arranged into sister chromatids. Since the aim is to have half the normal amount of DNA, a second phase of cell division occurs with the chromatids detaching and segregating as in mitosis: finally resulting in four germ cells being produced from a single stem cell, each of which is genetically different to all the others.
Recombination occurs at the end of metaphase when pairs of homologous sister chromatids have become aligned at the cellular equator. This alignment is called synapsis. Crossover is the swapping of genetic material between chromatids bought into close contact by synapsis. A number of points are randomly selected along the length of the two chromatids and sections of the DNA occurring between every other pair of points are swapped. Compared to the length of the chromatids, these crossover points are in-numerous and are most likely to occur within regions of non-coding DNA. Therefore, the effect is that genes, or groups of genes, move between pairs of chromosomes. Most of the time, the integrity of genes are unaffected. Sometimes a crossover point will occur within a gene and, most likely, this will result in a novel gene formed from parts of the paternal and maternal versions. However, since chromosomes do not always align perfectly, there is some chance of this new gene not working correctly.
The Holliday model was the first to offer a satisfactory explanation of the mechanism behind crossover. At synapsis, both chromosomal duplexes align. Under the control of enzymes, a nick is made in a single DNA strand in one of the chromosomes. At this point, a small section of the nicked strand becomes detached from its complementary strand and invades the other chromosome, which is nicked at the same point, ligating to the corresponding strand. The same effect happens with the displaced strand from the other chromosome, forming a crossed-strand Holliday structure. This crossed section linking the chromosomes is mobile, able to move in either direction along the DNA complex. Under enzymatic influence, the crossed section moves a random distance - a process called branch migration - further detaching sections of single-strand DNA and carrying them over to the other chromosome. Rotation around the crossed section, the crossover point, results in an isomeric Holliday structure, a cross-shaped formation characteristic of recombination. Further nicks and resealings, driven by enzymatic action, produce one of two different end structures - recombinative or non-recombinative. Both of these contain heteroduplexes, sections of non-complementary duplex DNA. These will either be repaired by the DNA excision-repair mechanism, or remain heteroduplex until the next cell division. At this point, each daughter chromosome will gain a different section of DNA corresponding to the two halves of the heteroduplex. In recombinant DNA, sections of duplex DNA have transferred between chromosomes. Heteroduplexes occur around the joining point. In non-recombinative DNA, only small sections of single-strand DNA have been transferred between chromosomes. The excision-repair mechanism can update from either the existing strand, replacing the transferred single-strand, or from the invading strand. If crossover occurs within a gene, this latter action will cause gene conversion, the updating of an exon with new code.
1Other factors deciding the final shape include the nature of the peptide bonds - which behave like inflexible double bonds due to resonance from the nearby C=O bond - and the possible presence of prosthetic groups (see later).
2Recognition here, and elsewhere in molecular systems, refers to a stable non-covalent bonding between large macromolecules - a result of diffusion rather than active convergence.
3A week form of non-covalent bonding whose net effect can be quite appreciable between large complementary-shaped molecules.
4This proximity suggests that similar genes may be a result of unequal crossover - crossover between chromosomes which are not properly aligned.
5Which gives mobile DNA the alternative name of selfish DNA. This should not be confused with Dawkins' idea of the selfish gene, which is unrelated.
6During interphase, chromosomes remain associated with this scaffold. Binding sites between the chromosome and scaffold are called scaffold-associated regions (SAR's), occuring between transcription regions. During transcription, the solenoid in the transcription region unwinds to allow access by transcription proteins.
7Telomeres consist entirely of guanine bases, forming a loop at the end of the chromosome. The sides of the loop are bound together by non Watson-Crick G-G bonds.
8This DNA is passed, through mitochondria in the female egg cell, from a mother to her offspring. It is unrelated to nuclear DNA and, since since sperm do not contain mitochondria, does not recombine with male mitochondrial DNA. Mitochondria are thought to be prokaryotes that formed a symbiotic relationship with the eukaryotic cell. (This also applies to chloroplasts).
9The CG dinucleotide is highly susceptible to mutation, whereby methylation causes C to mutate into T. This explains its under-representation. A special DNA repair system protects those stretches of CpG used as promoters.
10These regulation proteins are able to bend DNA. During transcription activation, this allows distant proteins to move close together, causing large loops of DNA to appear.
11Sexually reproducing bacteria do so
by horizontal gene transfer, whereby a section of DNA from one bacteria invades the DNA
of another. This can occur whenever the bacteria are in proximity.