XSPAN
User Interface
The
GUI allows users to select a target organism or cell
ontologyfrom a drop-down
list. The anatomy of the selected organism/ontology
appears in the left hand display window. The
user then browses the anatomy displayed and
highlights the initial tissue to be used for searching.
A
target organism (or "All") is selected from
the second drop-down list and this anatomy/ontology
diplays in the right
hand display window. A mapping type (or "All")
is selected to determine the basis of any tissue-to-tissue
mappings.
If "Expert" or "Derived" are chosen,
a mapping sub-type drop-down list appears to allow the
user to refine the
mapping type.
The
user then clicks the Search button and any mappings
between the selected initial tissue and the target organism/s
are retrieved from the database. These are listed according
to target organism in the Results Table.
Clicking
on a row in the Results Table will bring up the target
organism for that row in the right-hand display, and the
mapped tissues are highlighted with the tree expanded
to the mapped tissues. An new frame with full details of
the mapping then appears to the right.
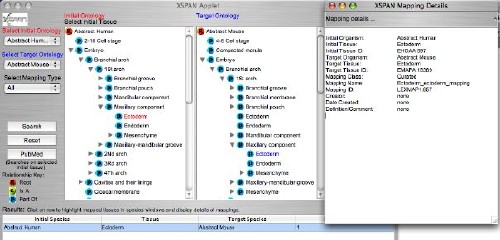
Screenshot
of the XSPAN applet GUI.
Click here or
on the image above to see a larger version of the screenshot.
The XSPAN GUI (Graphical User Interfaces) is now publically available, running as an applet. Click on the link below to access it.
XSPAN applet
To access the Help Pages for the Applets click on the following links:
XSPAN Applet Help
Click
on a menu option for more details of XSPAN. |



