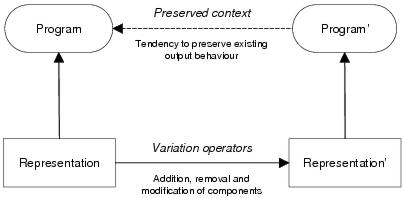
Figure 10.1: The combination of implicit context and variation filtering causes a tendency to preserve existing context and program output behaviour.
This chapter summarises the work reported in this thesis and the rationale behind it; presents conclusions; discusses limitations of the experimental method; and speculates about further work.
This thesis began with the goal of mining biological knowledge to improve the behaviour and performance of genetic programming. Biology has served as an inspiration to computer science throughout its history. Many fields of modern computer science, such as machine vision, robotics, and neural networks, place an implicit emphasis upon the mimicry of biological systems in both their goals and their methods. Indeed, biology possesses many of the attributes which many people would one day like to see in computer systems, including: fault tolerance, self repair, autonomous behaviour, self-replication, intelligence, learning, reconfigurability and environmental interaction. Yet, according to Charles Darwin's theory of evolution, all these attributes came about through an iterative process of selection acting upon random change to some initially very simple organism - and herein lies the motivation behind evolutionary computation.
Nevertheless, both our understanding of biology and our understanding of change within non-biological systems points to the realisation that a complex artifact can only be evolved if the artifact is internally organised in a suitable way. For example, it is very difficult to derive a complex software system from a basic software system if the basic software system does not have a meaningful architecture or is not written in legible code - and this is true even if the changes are being made by an experienced and intelligent programmer. Within biological systems, genetic change is essentially random: yet biological systems are still considered to be highly evolvable. This is because biological systems are internally organised in a way that, to an extent, prevents inappropriate change and promotes meaningful change. The sources of evolvability within biological systems exist at many levels and include: redundancy, compartmentalisation, exploratory mechanisms, and epigenetic and ecological processes. However, perhaps one of the most important sources of evolvability in biology is that behaviour emerges bottom-up from interactions between biological components which implicitly know what they want to do and what they want to do it to. This characteristic has been termed `implicit context' within this thesis.
By comparison, most genetic programming systems use what could be termed `explicit context' representations: where the interactions between components are determined explicitly by their position within the representation. However, such positionally dependent representations have important limitations: (i) a component loses its context, and hence changes its behaviour, if its relative position is changed; and (ii) particular positions can have different meanings within different programs. This implies that recombination operators, which move components between programs and typically change their position, rarely preserve the behaviour of the components they recombine; with the consequence that co-operative evolution - a conceptually powerful form of evolution - can not easily occur within genetic programming populations.
A number of non-standard GP systems [Poli, 1997,Luke et al., 1999,Miller and Thomson, 2000,Oltean and Dumitrescu, 2002] use an alternative form of representation which, in this thesis, is termed `indirect context'. In an indirect context representation, each component is assigned a reference and each component specifies its interactions with other components according to these references. Explicit context can be seen as a special case of indirect context in which a component's reference is its position within a program representation and in which components can only occupy positions which are referred to by other components. In an indirect context representation, by comparison, components may be assigned references that are not referred to by other components within the representation. Consequently, these components are not used within the program described by the representation but can be used to support modes of evolution believed to be significant sources of evolvability in biological systems: such as neutral evolution and analogues of evolution be gene duplication. It is also possible for a component to be referred to by more than one other component, a condition comparable to pleiotropy in biological systems. Importantly, a component's reference need not be related to its position within the representation, and hence components can be moved within a representation without changing their behaviour. However, particular references can still have different meanings within different programs: and consequently indirect context does not prevent the loss of a component's meaning following recombination.
The work reported in this thesis concerns the development of an implicit context representation for genetic programming. The potential benefits of implicit context for evolutionary computation are very significant. Within biological systems, the behaviour of a biological component - both what it does and what it interacts with - is determined solely by its chemical constitution and physical shape: both of which are a consequence of its genetic definition. Loosely speaking, if during recombination a biological component's genetic definition is moved from the genome of one organism to the genome of another and the component becomes expressed, it will implicitly attempt to carry out the same behaviour in this new organism that it did in the previous organism i.e. its meaning will be preserved following recombination. This happens because biological components interact with other biological components according to recognition of one another's shape, which describes their behaviour, rather than one another's genomic position or other arbitrary reference. For example, the shape of an enzyme's binding site is complementary to that of its substrate; so the enzyme is effectively describing (in part) the behaviour of the component it would like to interact with.
The notion of an implicit context representation for genetic programming is based upon this biological concept of implicit context. Rather than program components specifying their context via explicit position or indirect reference, in an implicit context representation components specify their interactions in terms of the behaviour of the components they would like to interact with. However, in order for this to be possible, components must be able to interrogate other components to determine their behaviour. Assume for the moment that there is a perfect mechanism whereby components can determine the behaviour of others. Following recombination, a component would be able to inspect other components and determine whether or not there are any components it should interact with in order to achieve its pre-defined behaviour. Note that there is no requirement for a component to become involved in a program if there are no suitable interactions. In this situation the component would become recessive. However, any behaviour which a component does carry out will be consistent across programs.
Variation operators can alter a program representation in four different ways. They can add components, remove components, re-order components, or change (mutate) the behaviour of components: either by changing how they should interact with other components (their function) or by changing which components they should interact with. The use of implicit context leads to a phenomenon which is termed `variation filtering' within this thesis; whereby particular variation events can be wholly or partially ignored. For example, if a component is added to a program representation, then it will only affect the program's outputs if it interacts with existing components that are expressed in the program i.e. only if it fits into the existing contexts declared by the program. If an expressed program component is removed from a program representation, then it might be possible for the other expressed components to compensate for its absence by interacting with a previously recessive component with a similar behaviour. This could also happen if an expressed component lost its previous behaviour as a result of a mutation event; or conversely a mutation event might cause a previously recessive component to offer a better match to an existing component's interaction preference than a currently expressed component. In effect, there is a tendency for variation events to be ignored or partially compensated for if they cause change which does not fit the contexts currently declared by the program. This, in turn, promotes gradual change in program behaviour: something which is conceptually desirable from the perspective of evolvability.
Enzyme genetic programming is a form of genetic programming based upon an implementation of implicit context representation. In enzyme GP, a program is a collection of inter-connected functions, program inputs, and program outputs. A program is represented by a linear array of program components, each of which has: an activity; a behavioural description; and - if it processes input - a set of input preferences, which describe the behaviour of the components whose outputs it would most like to receive inputs from. A program representation is mapped into a program via a development process which attempts to connect the inputs of every component to the outputs of those components which most closely match their input preferences.
Unlike in biology, it would be insufficiently precise to describe a program component by its activity alone, since both within a single program and within a population of programs there will be many program components with the same activity but with different roles. To overcome this problem, a program component, in enzyme GP, is described by both its own activity and the activities of the components which develop below its inputs. However, the exact components which develop below a component's inputs can not be known until after the development process is complete. Consequently, enzyme GP must describe a component's behaviour in terms of its expected inputs; those components which would be connected to its inputs if its behavioural preferences were matched precisely during development. This description is called a functionality and, in effect, declares a component's behaviour as an expected activity profile, weighted by depth, of the components that occur in the program fragment of which it is the root.
Enzyme genetic programming has been applied to a range of discrete symbolic regression problems. Analysis of the performance and behaviour of the evolutionary process has led to a number of conclusions.
Meaningful variation filtering is defined as the capacity for an evolutionary system to filter out inappropriate change caused by the action of variation operators. Theoretically (see above), the use of implicit context representation should allow an evolutionary system to carry out meaningful variation filtering. This is supported by a significant amount of experimental evidence indicating that enzyme genetic programming does carry out meaningful variation filtering.
The main experimental evidence that enzyme GP carries out meaningful variation filtering lies in the ability of enzyme GP to carry out meaningful recombination. When compared to indirect context, enzyme GP's implicit context representation leads to a substantial improvement in recombinative performance: both in the capacity for a program representation to receive components transferred from another program representation, and in its capacity to preserve the meaning of the transferred components. These quantitative results are supported by the identification of context-preserving behaviours during qualitative analysis of the effect of recombination upon individual programs.
It has been hypothesised that the redundant portions of implicit context program representations are organised into a subsumption hierarchy; and that this subsumption hierarchy supports meaningful variation filtering by absorbing inappropriate change and saving subsumed program components in case they are needed for back-tracking. The utility of this structure can be seen in the observation that the non-coding portion of representations grows in response to the addition of components; filtering out components which do not fit into the contexts defined by programs. Evolutionary performance is substantially degraded when redundancy is stripped from program representations following program development. This supports the views of other researchers that functional redundancy is an important source of evolvability within evolutionary representations [e.g. Smith and Goldberg, 1992,Dasgupta and McGregor, 1993,Haynes, 1996, Levenick, 1999,Yu and Miller, 2002 ].

Figure 10.1: The combination of implicit context and variation filtering causes a tendency to preserve existing context and program output behaviour.
Output terminal components have an important role in program development for they uniquely determine the program's output context and therefore which components will contribute to the program behaviour following development. Furthermore, by attempting to make the same input connections, they attempt to implement the same output behaviour irrespective of which components are actually present within the program representation. Consequently, so long as the output terminal components remain the same, they should function as an attractor, tending to preserve the existing program output behaviour irrespective of changes that occur within the program representation as a result of the application of variation operators (this idea is depicted in figure 10.1). This, within enzyme GP, should be the ultimate effect of the variation filtering process enabled by implicit context representation. This effect should also occur where recombination leads to a child program containing output terminal components from more than one parent program. In this case, there will be a tendency to preserve the output behaviour from each parent for the corresponding output terminal: which, in many cases, is exactly the behaviour that is wanted from recombination.
This is suggested by the considerable positive effect of a low level of Lamarckian reinforcement learning; where program components' input preferences are changed so that they more accurately reflect the input connections they have made during development. The effect of this is to strengthen the internal cohesion of a program over time, increasing the effect of variation filtering and thereby making it less likely that large changes will occur as a result of variation events. This supports both previous results on the utility of Lamarckian reinforcement learning [Teller, 1999,Downing, 2001], and the view that compartmentalisation can be an important source of evolvability [Conrad, 1990,Kirschner and Gerhart, 1998].
It is difficult to make general conclusions regarding the effect of implicit context representation upon the evolution of program size, since program size within enzyme GP appears to be significantly affected by inherent biases within the functionality implementation of implicit context. However, it can be speculatively suggested that GP with implicit context representation should suffer significantly less from bloat than GP with explicit context representation, given that: (i) recombination is less disruptive; (ii) the recombination mechanism does not promote the development of protective syntactic introns; (iii) the effect of removal bias is far smaller; and (iv) program size is decoupled from representation length. This argument is somewhat supported by the observation that there is little or no bloat in enzyme GP. It has also been hypothesised that small programs are better replicators than large programs, since they have less internal connections and are therefore less likely to be disrupted by variation events. This is also supported by the tendency towards the evolution of small programs in enzyme GP. However, there is some concern that this might make it harder to solve problems which have large optimal solutions and relatively short sub-optimal solutions - although this concern is somewhat allayed by the diversity preservation mechanism that is inherent in implicit context representations.
These conclusions support the hypothesis that models of biological representations can be used to represent computer programs and other artificial executable structures within genetic programming, thereby improving the evolvability of these structures.
Whilst considerable effort has been made to verify these conclusions through both experimental investigation and theoretical understanding, there are limitations of the experimental method which might reduce their generality. The most significant limitation is that the performance of enzyme genetic programming has not been vigorously analysed upon a wider class of problems; and whilst discrete symbolic regression was chosen for its relative difficulty with regard to recombination, it can not be representative of all other problem domains. Partly this limitation reflects a need to generalise the functionality implementation of implicit context to handle so-called ephemeral random constants and structural behaviours in addition to functional behaviours. Suggestions as to how this might be done are given in the further work section. Furthermore, there has been little direct comparison between the performance of enzyme genetic programming and conventional genetic programming. This is due both to the lack of existing results applying conventional GP to Boolean regression problems, and to the difficulty of direct performance comparison caused by the use of a non-standard evolutionary framework within enzyme GP. However, it is assumed that explicit context representations have worse performance than indirect context representations upon recombinative search (for reasons given elsewhere in this thesis); and that therefore the performance of explicit and implicit context are being compared by transitivity. Nevertheless, it is hoped that these limitations will be addressed in future research.
Section 9.7.2 discussed how biases within functionality space may lead to size and structural biases during search. An important avenue for future work will be to look into ways of removing this bias. Bias can be reduced by warping functionality space so that changes within over-represented regions have less influence upon the evolutionary process. However, early work concerned with variable dimension lengths had led to little success. Perhaps a more effective approach, in the short term, will be to look at ways of biasing the initialisation and mutation mechanisms in order to counteract the biases in fuctionality space; although a more long term strategy should be to look at alternative reference spaces which do not have the same kinds of biases which are seen in functionality space.
Perhaps a more important short term goal should be to reduce the time complexity of development. The most obvious way of doing this would be to remove the non-recurrency constraint during development. Without this constraint, development will simply be a process of comparing the input preferences and behavioural descriptions of each pair of components; a process which will have far less complexity than maintaining and interrogating connection information at each node in order to determine whether or not a future connection will lead to recurrency. It will also allow the expression of recurrent structures: an important motif in many problem domains, including programming. If non-recurrent structures are still required, this constraint can be handled during execution by only propagating the first argument that arrives at a component output. However, it will be possible to evolve invalid structures in which there are missing paths between outputs and inputs; though if infrequent, these structures can be filtered out during selection. Furthermore, using the current approach to calculating functionalities, it may not be possible to represent recurrent structures exactly, since it may no longer be meaningful to give a profile of the sub-structure which develops below a component, given that the component may occur within this sub-structure via a recurrent connection. Nevertheless (as figure 10.2, discussed below, shows), it is still possible to give a profile of the activities which occur within the neighbourhood of a component. In any case, this will make little difference to program evolution, since actual evolved functionalities are unlikely to exactly describe components anyway.
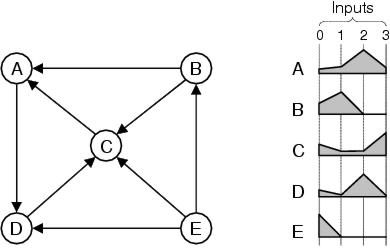
Figure 10.2: Implicit structural context. Shapes (right) describe the structural profile, weighted by distance, of each node in the structure on the left in terms of the degree of input connectivity within its local sub-graph.
In the long term, one of the most interesting avenues of research will be to look at forms of implicit context representation which are able to represent a far wider selection of structures. However, it will also be interesting to look at the potential of the current approach to implicit context. Within enzyme GP, functionalities attempt to describe profiles of the functional activities which occur within evolved structures. This approach can be generalised by describing profiles of `behavioural properties' within structures, which could include both functional and non-functional properties of their components. Figure 10.2, for example, shows how descriptive shapes can be used to represent an arbitrary digraph. Rather than describing the functions of the nodes within the structure, these shapes describe nodes by the number of incoming connections they have from other nodes. In theory, this approach could be used for any kind of structure; although in practice it would require that structures have sufficient heterogeneity in the connectivity of their nodes and the distribution of connectivity within the structure. However, as problems such as the two-bit multiplier show, the behavioural properties (e.g. activity types) within a structure do not have to be very diverse in order for it to be expressed and evolved by this method. Furthermore, there is no reason why a descriptive shape could not describe both functional and non-functional properties together. For example, the behaviours of components within digital circuits could be described by profiles of both the functional and structural properties of their expected sub-trees. There is also scope for describing continuous parameters. For example, ephemeral random constants have two functional properties: they are a constant input source and they provide a particular value. One functionality dimension could be used to describe the relative occurrence of constants within a program fragment and one could could be used to describe the relative value of the constants.
Recently, there has been some interest in using implicit context27 within manual programming environments. In an approach described by Walker and Murphy [2000], components analyse their call history in order to learn more about the behaviour they are expected to provide to other components. Because of this, components are able to request behaviour from one another via fewer function calls and fewer parameters; simplifying component interaction and leading to communication along narrow, well-defined paths. Due to cleaner interfaces and less complex connectivity, this form of implicit context, in principle, makes it easier for components to be re-used within other programs. Whilst only partially related to the idea of implicit context introduced in this thesis, this idea of a component learning its own context could be used within an EC implicit context representation. Lamarckian reinforcement learning is an existing mechanism whereby context learning can take place in the functionality model of implicit context. However, this form of learning is currently carried out at a program level and can only learn simple information about those activities which have dimensions within functionality space. A more complex form of learnt implicit context can be imagined in which a program consists of software agents which can flexibly learn about their environment and preserve this information when transferred between programs; gradually building up knowledge about the space of possible component interactions and their expected behaviour within this space. At the start of evolution, these agents would only have a general idea of the role they should fulfill - making behavioural exploration easy - but, during the course of evolution, they would become more specialised - improving behavioural exploitation.
Such behaviourally complex forms of implicit context representation could also be used within software engineering environments. Rather than programmers scripting the interactions between program components; implicit context would allow them to focus on writing components. When these components are placed in a program together, the program behaviour would emerge from implicit interactions between components. This would amount to applying biological principles of organisation to the realm of human design. However, this vision is currently rather far-fetched (and perhaps even impractical!). Perhaps a more productive goal in the meantime would be to apply more of the principles of biological organisation to the development of evolutionary computation representations. Thus far, in this thesis, this approach has led to the development of a representation modelled upon the interactions within metabolic pathways. It seems conceivable that this model could be extended to include the interactions between metabolic pathways and signalling and gene expression pathways. Gene expression pathways, in biology, are evidently the source of many of the more complex behaviours seen within biological systems. Signalling pathways are the predominant source of distributed processing within biological systems; and distributed processing appears to be an important factor both behaviourally and as a source of evolvability within biological representations. Certainly it is conceivable that the enzyme GP model could be extended to allow regulation of component expression by currently expressed components (a behaviour which could be particularly useful for solving dynamic problems) and to allow communication between `cellular' groups of components (which might themselves result from differential component expression). However, whilst making the system far more expressive, this will also make it harder to interpret by a human.
In addition to using enzyme GP as a mechanism for applying current biological understanding, it seems that enzyme GP, and other EC models of biology, could be used to discover biological knowledge. Certainly this has proved a productive approach for neuro-computing; which has helped add to biological understanding by testing the validity of neural theories, by allowing observation of animated neural models, by posing questions in response to implementation problems, and by testing the generality of neural processing through application to non-biological problems. Biological evolution is difficult to study using conventional data-centric bioinformatics techniques due to the sparseness of the fossil record and the slow pace of evolution. Evolutionary computation, by comparison, is relatively fast, can provide a complete `fossil record' and, in principle, could be used to to model, observe, pose questions about and study the generality of biological evolution. Perhaps the most beneficial use of this technique would be in understanding biological sources of evolvability; many of which can be modelled without reference to biological information. Existing EC research has already given some insight into the roles of redundancy and compartmentalisation - and there are still plenty of biological motifs, mechanisms, and structures which are not well understood in the context of evolvability. In particular, it would be interesting to consider whether enzyme GP might have a role in understanding biological evolution; particularly the early stages of biological evolution. It seems evident that variation filtering behaviours do occur in biological systems, although the relatively simple behaviours of the kind described in this thesis may only have had an influence before active forms of genetic and enzymatic regulation evolved. Likewise, it seems plausible that the non-coding portions of DNA have a role similar to the recessive subsumption hierarchy described in this thesis - providing a source of error recovery and evolutionary back-tracking.
27This use of the
term implicit context, whilst related, was coined independently.