Research Grants
Human Gut Cell Atlas – Normal Intestine and Crohn’s Disease: 2020-2023, Helmsley Charitable Trust-funded, Principal Investigator for Heriot-Watt University. (Lead-PI for Project Consortium: Prof Mark Arends, Edinburgh University)
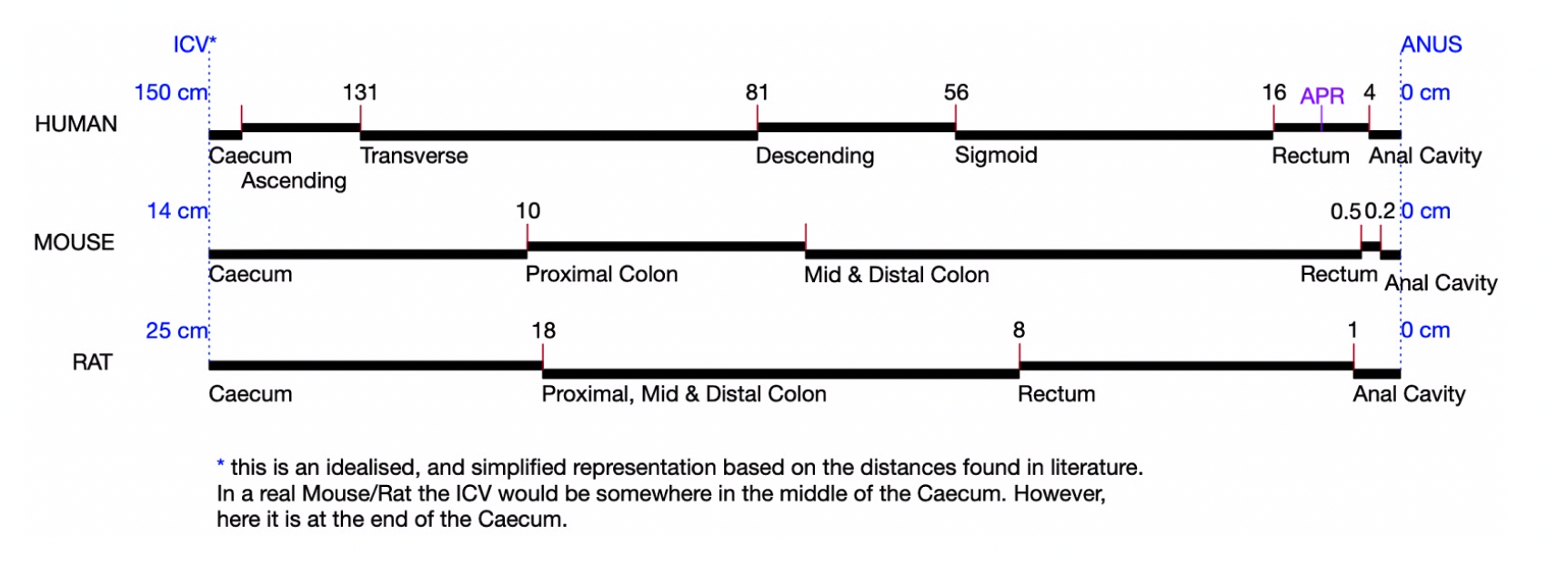
Summary: The goal of this study is to provide an integrated dataset of single-cell gene expression data mapped onto a high-resolution 3-dimensional framework model of the small and large intestines, focussing particularly on the terminal ileum, caecum and ascending colon. There will be mapping to both a 2D model (anatomogram) and a 1D linear model of healthy gut. Normal gut samples will be collected from appropriate subjects, as will samples from Crohn’s disease patients undergoing intestinal resections for complex fibrostenotic stricturing disease.
This project will also provide a gut-centric coordinate framework to map anatomical locations of cells providing novel insights into Crohn’s disease mechanisms, crucial to developing personalized therapeutic strategies, and new analytical tools for querying the Gut Cell Atlas database.
The project is a collaboration between:
- Pathology Division of the University of Edinburgh (Prof Mark Arends, Lead-PI, and Prof Richard Baldock)
- Department of Computer Science, Heriot-Watt University (Prof Albert Burger)
- Gene Expression group of the European Bioinformatics Institute (EBI), (Dr Irene Papatheodorou)
- The Wellcome Sanger Institute (Dr David Adams)
It is part of a wider GCA consortium funded by the Helmsley Charitable Trust. The Gut Cell Atlas is one of several organ-specific cell atlases developed under the umbrella of the Human Cell Atlas.
Comparative Workbench: 2018 – 2019, CZI-funded, Principal Investigator.
Summary: A collaboration between the Pathology Division of the University of Edinburgh (Prof Mark Arends, Lead-PI, and Prof Richard Baldock) and Heriot-Watt University (Prof Albert Burger, PI). The project consists of two main components: 1) the actual workbench tool (lead by the University of Edinburgh), a new software system for pathologists to organise images, typically microscopic images of varying types, along with a broader range of biological data, and 2) a model of human and rodent colons as a computational framework for spatially (anatomically) based integration of colon data (lead by Heriot-Watt University).
This is funded as part of the Chan Zuckerberg Initiative’s pledge to support the development of the Human Cell Atlas.
PhenoImageShare: 2013 – 2016, BBSRC-funded, Principal Investigator.
Summary: A collaboration between the MRC Human Genetics Unit (Prof. Richard Baldock, PI), the European Bioinformatics Institute (Dr. Helen Parkinson, PI) and the Biomedical Informatics Systems Engineering Lab (Prof. Albert Burger, PI), the PhenoImageShare project created a prototype infrastructure to support the sharing of phenotype annotations on phenotype images.
Further details: [PhenoImageShare Web Site]

Atlas Informatics: 2001 – 2012, MRC-funded, Biomedical Informatics Research Scientist, p/t secondment.
Summary: The Mouse Atlas project at MRC HGU provides a spatio-temporal framework for genomic and proteomic data about the developing mouse embryo. It consists of EMAP, the actual 3D mouse atlas and anatomy ontology, and EMAGE, a database of in-situ gene expression data mapped onto the atlas. As part of my work at the MRC I was involved in research and development of interoperability solutions for the Mouse Atlas.
Further Details: [Mouse Atlas Project Web Site]
CUBIST: Combining and Uniting Business Intelligence with Semantic Technologies: 2010 – 2013, ICT EU-funded, Principal Investigator
Summary: The goal of CUBIST was to develop new Business Intelligence (BI) solutions, specifically through the application of Semantic Web technologies, to address the challenge of every increasing amounts of non-structured data; traditionally, BI only deals with structured data. At Heriot-Watt, we focus on a biomedical use case for CUBIST, in particular the semantic representation of image data in the Mouse Atlas and its exploitation through the CUBIST system.
Further Details: [CUBIST on BISEL pages]
RICORDO: Interoperable Anatomy and Physiology: 2010 – 2013, EU-funded (Virtual Physiological Human), Principal Investigator
Summary: The RICORDO project focused on the study and design of a multiscale ontological framework in support of the Virtual Physiological Human community to improve the interoperability amongst its Data and Modelling resources. To this end, it aimed to build directly upon the shared experiences and published recommendations emerging from the VPH Network of Excellence and ELIXIR initiatives.
Further Details: [RICORDO on BISEL pages]
INCF: Digital Atlasing Program: 2008-2010, INCF-funded; Task Force Member and Principal Investigator for Pilot Study (4/09-10/09).
Summary: Digital atlases have been identified as potential neuroinformatics frameworks, as they act as a two-dimensional or three-dimensional map that one may use to traverse the brain and associated data of different granularity, modalities, and sources. The Task Force investigated the state of the art of rodent brain digital atlasing and formulated standards, guidelines, and policy recommendations.
A spin-off from this activity was a pilot project investigating the integration between the Edinburgh Mouse Atlas Project (EMAP) and the Allen Brain Atlas (ABA). Specifically, in BISEL we explored the use of the NIF integration infrastructure. We were also involved in the development of a INCF hub for the Edinburgh Mouse Atlas Project as part of the INCF Distributed Atlas Infrastructure (INCF-DAI).
Further Details: [INCF on BISEL pages] [INCF web site]
Argudas: An Argumentation System for Gene Expression Databases: 2009 – 2011, BBSRC-funded; Principal Investigator;
Summary: The objective of the Argudas project was the development and deployment of a web-based argumentation tool that allows biologists to discover and assess inconsistencies and incompleteness within and across multiple gene expression databases. It presents the user with arguments for and against specific gene expression statements and, if requested by the user, will weigh up the evidence from the different databases to suggest an overall conclusion with respect to whether or not a particular gene is expressed in a given anatomical structure.
Further Details: [Argudas on BISEL pages]
Sealife: A Semantic Grid Browser for the Life Sciences: 2006 – 2009, ICT EU-funded; Principal Investigator;
Summary: The objective of Sealife was the conception and realisation of a semantic Grid browser for the Life Sciences, which links the existing Web to the currently emerging eScience infrastructure. At Heriot-Watt, we developed solutions for the automatic composition of web services, using AI planning, applied to gene expression databases. Our work on Argumentation Systems in the Life Sciences also originated as part of Sealife.
Further Details: [Sealife on BISEL pages]
REWERSE: REasoning on the WEb with Rules and SEmantics: 2004 – 2008, EU-funded, Prinicipal Investigator;
Summary: REWERSE was a research “Network of Excellence” (NoE) on “Reasoning on the Web”. The project addressed the IST strategic action line “Semantic-based knowledge systems”, aiming to establish Europe as a leader in reasoning languages for the Web.
REWERSE involved 27 European research and industry organisations from 14 European countries, including about 100 computer science researchers and professionals, co-ordinated by the University of Munich.
Further Details: [REWERSE on BISEL pages] [REWERSE project web site]
XSPAN: Cross Species Anatomy Network: 2002 – 2005, BBSRC-funded, Principal Investigator;
Summary: XSPAN was a collaborative project with the University of Edinburgh and aimed to support cross-species access to tissue-based genetic information through an internet-based cross-species anatomy network, i.e. a cross-species anatomy ontology integration system. The system provides interoperability between databases of key model organisms which facilitates access to gene expression and other genomics and proteomics resources.
Further Details: [XSPAN on BISEL pages] [XSPAN project web site]
SCANWEB: Cancer Information Network: 2002 – 2005, NOF-funded, Co-Investigator (Principal Investigator from March 2005);
Summary: This was a collaborative project with Lothian Health Board to develop online resources for the South East Scotland Cancer Network.
Further Details: [SCAN on BISEL pages] [SCAN project web site]
Other earlier projects:
- genISYS: Visualising Complex Datasets in Clinical Cancer Genetics (co-investigator; Scottish Office funded;
- Data Warehousing & Data Mining to support Internet Bureau Service, Findlay Irvine (co-investigator, DTI funded, www.findlayirvine.com – Company Web Site);
- Distributed Interactive Simulations (co-investigator, DERA funded);
- Performance Analysis of Parallel Databases (joint project when joining Heriot-Watt University, EU and EPSRC funded);