BISEL
Biomedical Informatics Systems Engineering Laboratory
Argumentation
Sealife Home | TCM | GGAPS | Argumentation | Evaluation | Publications
Biologists publish experimental results online. For various reasons these experiments can produce contradictory conclusions. Because the resources try to be as complete as possible, these results may be published. Consequently a single resource can contradict itself. As many fields have multiple online repositories, there is also the possibility of inconsistency between two, or more, databases. Such inconsistencies cause frustration for users, and can result in a workflow producing incorrect output. There are a range of possible solutions, however we chose to explore Argumentation.
We imagined Argumentation being implemented in an extra module that could sit alongside the existing SEALIFE components. This could be called into action if the user had any doubts over the output of a service run by the TCM. The final conclusion could then be added to the CART for later use by the TCM. The flow is summarised in the following diagram:
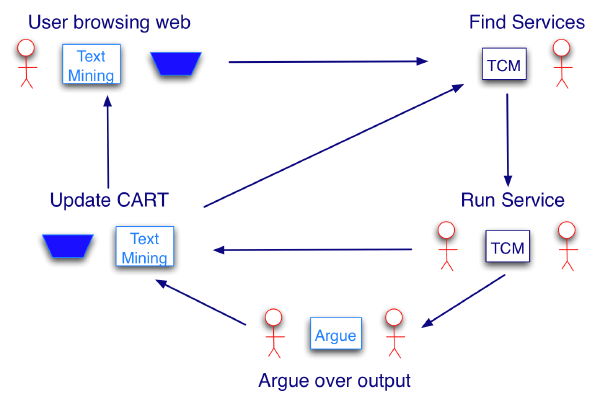
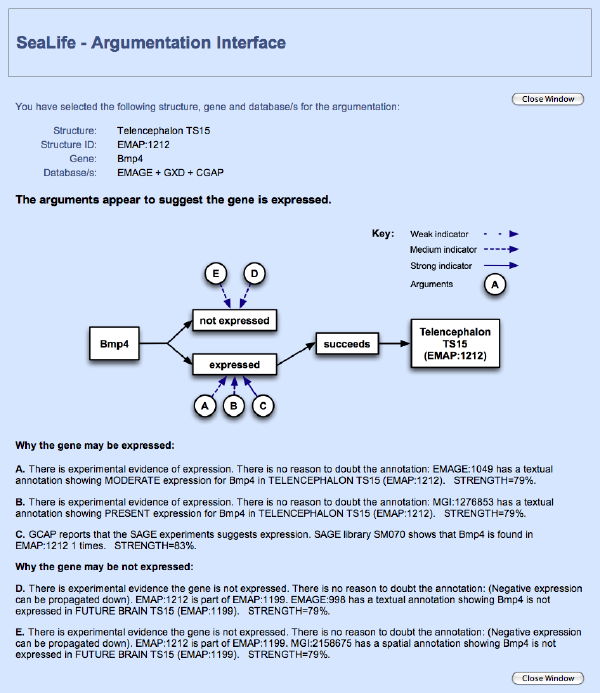
Argumentation is essentially the evaluation and balancing of arguments supporting a conclusion, against those attacking it. In this case arguments were created for, and against, a particular gene being expressed in a particular structure. The step step in this process, is the creation of argument template (called schemes) from which the actual arguments can be generated. These schemes were modelled on the reasoning of an expert user in our chosen domain (in-situ gene expression for the developmental mouse). Once the schemes were produced, they were used with a pre-existing argumentation engine (www.argumentation.org) to generate arguments.
This engine took as input the schemes - converted into inference rules - and domain information that was pulled from two appropriate data resources (EMAGE and GXD). The information allowed the variables in the rules to be instantiated, generating actual arguments that could be presented to the user. The presentation of these arguments was investigated as part of the evaluation for the SEALIFE project under which this work was done. SEALIFE Evaluation Website. Ultimately, it is the responsibility of the user to determine, based on the arguments, whether or not the gene is expressed.
Further details of this work can be found in the following publications:
Using Argumentation To Tackle Inconsistency And Incompleteness In Online Distributed Life Science Resources.
Paper for IADIS - International Conference Applied Computing 2007.
Kenneth McLeod and Albert Burger.
Download PDF
Download reference as BibTeX file
Towards the use of Argumentation in Bioinformatics: A Gene Expression Case Study.
Paper for ISMB 2008.
Kenneth McLeod and Albert Burger.
Download PDF
Download reference as BibTeX file
Click on the link below to visit the SEALIFE Project website:
http://www.biotec.tu-dresden.de/SEALIFE