BISEL
Biomedical Informatics Systems Engineering Laboratory
Evaluation
Sealife Home | TCM | GGAPS | Argumentation | Evaluation | Publications
Aim of Evaluation
The aim was to evaluate the usability of the Sealife systems prototype GUI and carry out summative and formative evaluation of each sub-system’s functionality and aspects of data presentation and visualisation for the systems. The evaluation was also used as a focus for expert user evaluation of the underlying system processes by select users.
User Group
Eighteen users were recruited, ten from the Edinburgh Mouse Atlas Project (EMAP) at the MRC in Edinburgh (in various roles from system developers to biological database curators) and eight Computer- and Life Sciences students at Heriot-Watt University. For the analysis users were divided into the two groups, nine Biologists and nine Non-biologists, based on background and experience. The Biologists included expert users who gave additional expert opinion on aspects of the system.
Procedure
A prototype GUI was designed and implemented and test scenarios designed, based on typical tasks the system is intended to perform. Data generated by the underlying engines was hard-coded into the GUI to forestall issues of availability and variation of third-party online resources during the course of the evaluation. Appropriate time delays were introduced.
Two observers conducted the evaluation, one interacting directly with the user, and the other observing the user and their interaction with the system, recording timings, errors, comments made by user and general observations on user actions. Users were prompted to comment, ask questions or ask for help freely at any stage during the evaluation.
Protocols
The protocols used for the evaluations are attached as appendices to the Technical Report (click here to view). The general usability questions were adapted from Shneiderman’s Questionnaire for User Interaction Satisfaction (QUIS). The same format was used where appropriate for the other questionnaires along with other more specific question formats.
Scenarios and Questions
The GGAPS evaluation consisted of two scenarios followed by questions specific to the GGAPS system. The first scenario required the user to step through a bioinformatics task using the browser-based user interfaces of appropriate online resources. The task involved discovering genes that might be expressed in the early stages of development in the human brain. The second scenario involved the user employing the GGAPS system via the GGAPS GUI to perform a similar task. Timings for each scenario were collected, with splits to allow for variable delays in return of online data to be accounted for. The user was asked to rate the usability of each system and specific questions relating to the presentation of information and data in the GGAPS GUI.
The Argumentation scenarios consisted of a walkthrough relating to expression of a gene in the developmental mouse brain, using the default settings. The user was then presented with a graphical and textual representation of the argument process and outcome. They were asked specific questions regarding the process and presentation of the results. The second scenario involved the user walking through the same process, but altering selections for some of the parameters. The users were then presented with the argument, modified appropriate to the altered parameters, and again asked specific questions regarding the results and their presentation. Users were also asked questions regarding their understanding of and views on argumentation and process of the argumentation system.
Results
The evaluation showed that the usability of the Sealife system as a whole is good. Below is a graphical representation comparing the over-all usability for both the GGAPS and Argumentation parts of the system. The ratings given by each user for each question have been added, to giving an overall impression of the distribution of ratings for the system.
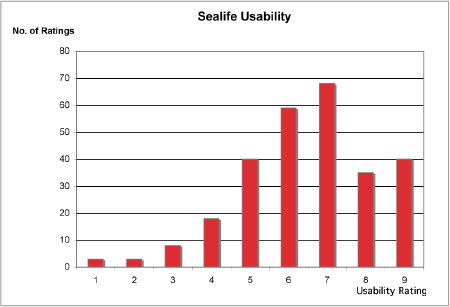 |
Click on image to see large version. |
GGAPS
The GGAPS system was favoured by the users over the manual approach for performing the evaluation scenario with statistically significant better ratings of the GGAPS system recorded. Users rated it as easier, quicker, more efficient, and easier to understand than the manual approach. The flipside of this was that the users felt they had less control using the system, but this may not be a significant issue as the majority of users trusted it to achieve their goal with no extra input from themselves.
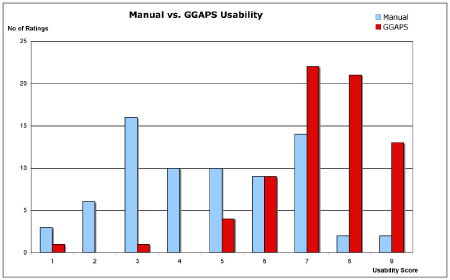 |
Click on image to see large version. |
There was a statistically significant time saving by using the GGAPS system over the manual system, and this could represent a considerable time saving in the course of a biologist’s work.
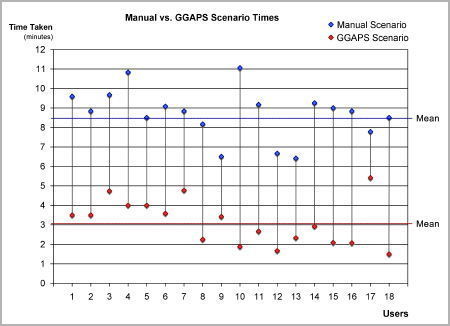 |
Click on image to see large version. |
A number of comments were recorded indicating that users wanted more detail of underlying data and processes, yet very few users examined the extra information the system provided such as viewing the expanded details of the proposed processes or the full BLAST report provided.
Valuable user feedback from comments and observations was gathered through the evaluation and this will inform design decisions for future versions of the GGAPS system. These will include addressing issues with user interaction, the presentation of results and visualisation of the underlying processes.
In summary, the evaluation showed that GGAPS represents an important step towards a system that could help biologists to achieve their goals more efficiently without requiring in-depth technical knowledge of web services. Such a system will also enable biologists to consider the use of previously unknown resources and realise potential goals they would not otherwise have considered.
Argumentation
The division of the user group into the Biologists and Non-biologists group was particularly pertinent to the analysis of the Argumentation System evaluation results. Within the Biologists group, four users were identified as "expert" in the field of gene expression. Detailed analysis of the answers and comments given by the Biologists (experts and non-experts), compared with the Non-biologist group enabled a number of important issues relating to the use of argumentation in biology to be examined.
The Argumentation system was well received by users and enabled both Biologists and Non-biologists to reach the correct conclusions regarding gene expression in the developing mouse. However the contrived nature of the second scenario raised issues of trust of the system by expert users.
Valuable insight and feedback was obtained on which elements of the presented results users employed in reaching conclusions. All elements were used to a greater or lesser extent by both groups of users, indicating the need for both graphical and textual representations to be presented.
Comments on the layout used for the graphical representations lead to an extension of the evaluation, using an online survey, to examine preferences for different graphical representations. The survey showed 62% of respondents preferred a Toulmin-like graph representation of an argument over a tree-like representation (examples below). However, when considering the tree-like representation, 81% of respondents preferred the top-down tree form over the bottom-up form, traditionally used in argumentation.
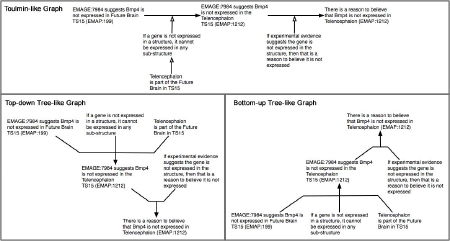 |
Click on image to see large version. |
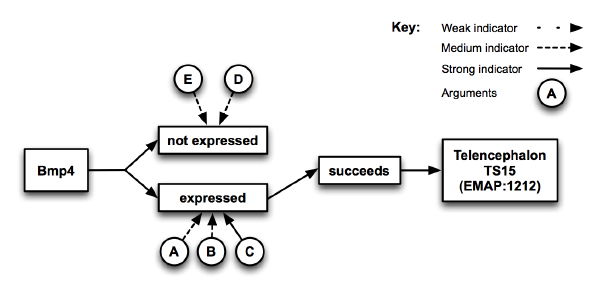
The visual technique used to summarise the arguments was tested to ensure it was understandable (example below). Users were correct 87% of times when asked to identify the strength (strong, moderate, or weak) arguments represented in the diagram. Apart from two outliers, most of the users found the diagram very easy to understand.
Conclusion
Valuable feedback on the user interface, usability, functionality and methods of data presentation was obtained through the evaluation process. This will form the basis for future developments of the system and also feed into work addressing the wider issues involved in developing systems to assist biologists in undertaking bioinformatics tasks.
A full report on the Sealife Evaluation is published at:
Sealife Evaluation.
G. Ferguson, K. McLeod, K. Sutherland and A. Burger.
Dept of Computer Science, Heriot-Watt University.
Internal Report Number = 0063.
2009.
Click here to view or download.
Click on the link below to visit the SEALIFE Project website:
http://www.biotec.tu-dresden.de/SEALIFE